Why proteins fold and how GPUs help us fold
Source: Hacker News
Before We Talk About AI, We Need to Talk About Why Proteins Are Ridiculously Complicated
You know what’s wild? Right now, as you’re reading this, there are approximately 20,000 different types of proteins working inside your body. Not 20,000 total proteins, 20,000 types. The actual number of protein molecules? Billions. Trillions if we’re counting across all your cells.
Each one has a specific job and a specific shape. If even one type folds wrong, it can lead to diseases such as Alzheimer’s, cystic fibrosis, sickle cell anemia, Parkinson’s, Huntington’s, mad cow disease, or any of thousands of other protein‑misfolding diseases.
Your body makes these proteins perfectly, billions of times a day, in every single one of your ~37 trillion cells—without a user manual or a workshop.
For decades, scientists tried to predict what shape a protein would fold into based solely on its amino‑acid sequence. Entire careers, Nobel Prizes, and supercomputers were devoted to this problem. Then AI companies arrived in 2020, claimed to have solved it, and indeed made rapid progress.
Now we’re not only predicting shapes; we’re designing entirely new proteins that can break down plastic, capture carbon dioxide, or target cancer cells with high precision.
But before we dive into NVIDIA’s role, let’s understand what proteins actually are and why folding is such a formidable challenge.
Proteins 101: The LEGO Bricks of Life (Except Way More Complicated and They Build Themselves)
Recall the central dogma: DNA → RNA → Protein. Information flows one way (retroviruses aside).
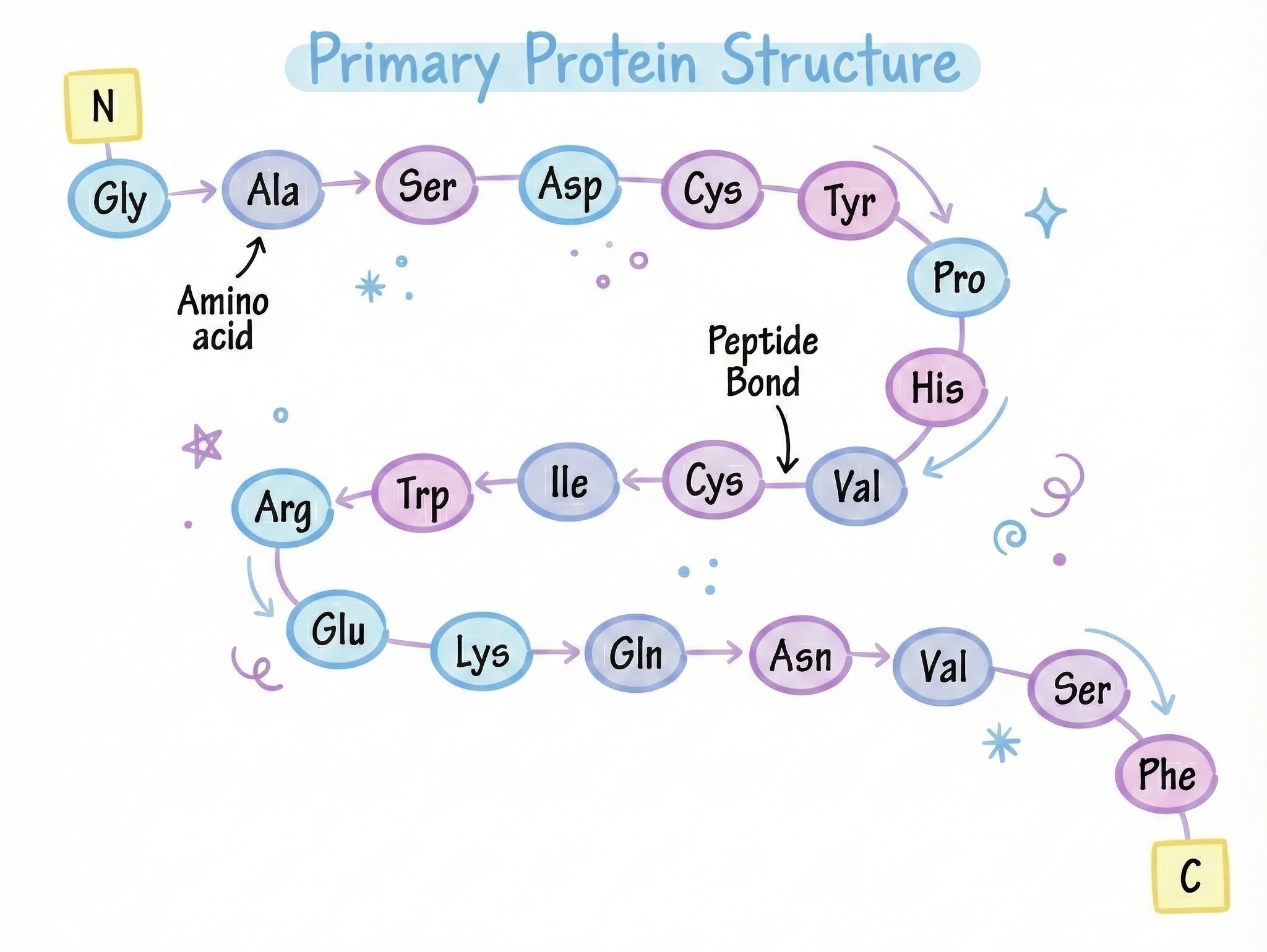
A protein is a chain of amino acids that folds into a specific 3D shape, and that shape determines what the protein does.
A chain → folds → shape → function.
Amino Acids: The 20‑Letter Alphabet That Writes Every Function in Your Body
There are 20 standard amino acids that your body uses to build proteins (a few non‑standard ones exist, but we’ll ignore them for now). Think of them as letters in an alphabet that assemble functional machines.
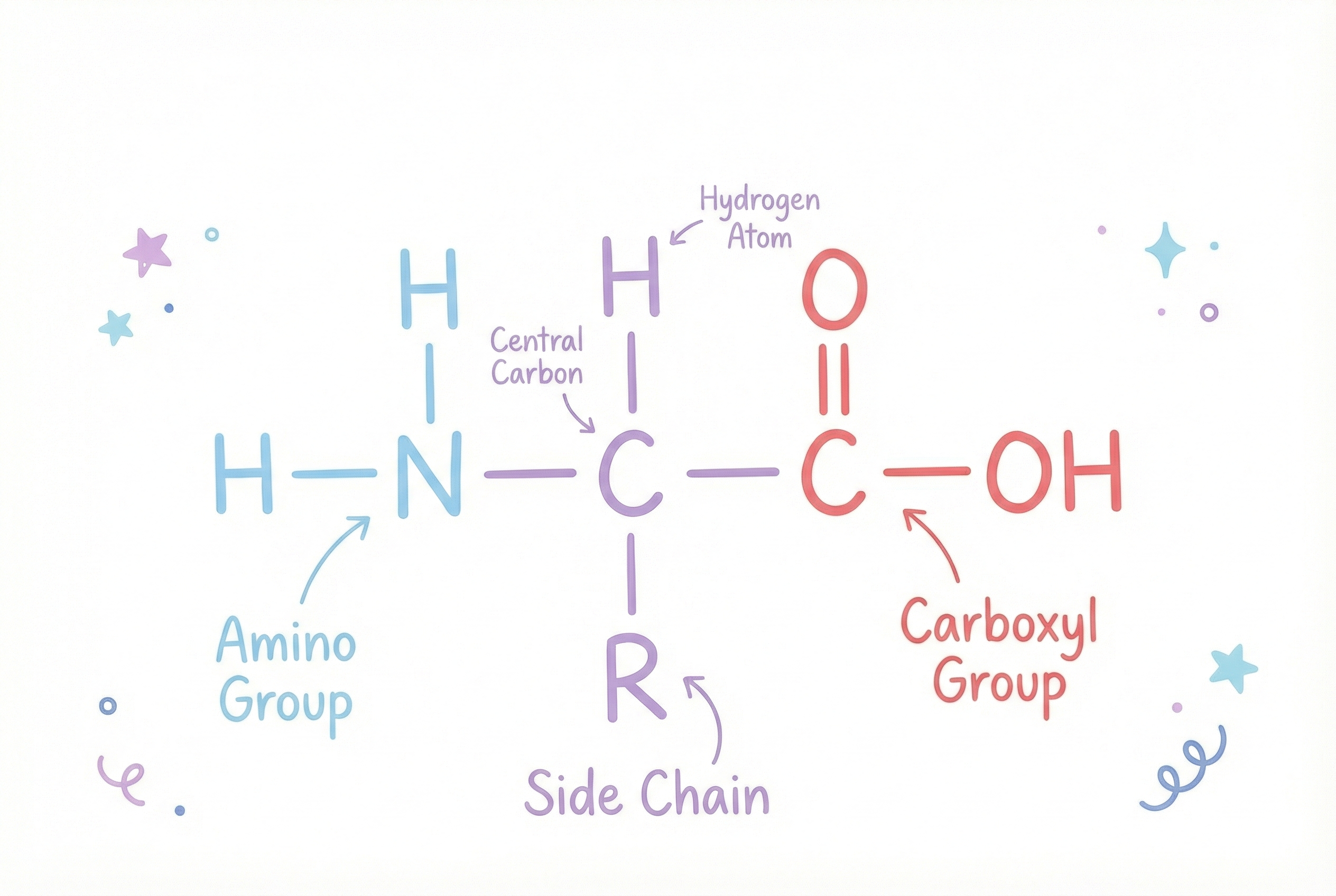
Each amino acid shares a common backbone:
- Amino group (NH₂)
- Carboxyl group (COOH)
- Hydrogen atom attached to the central carbon
…and a unique side chain (R group) that gives each amino acid its personality.
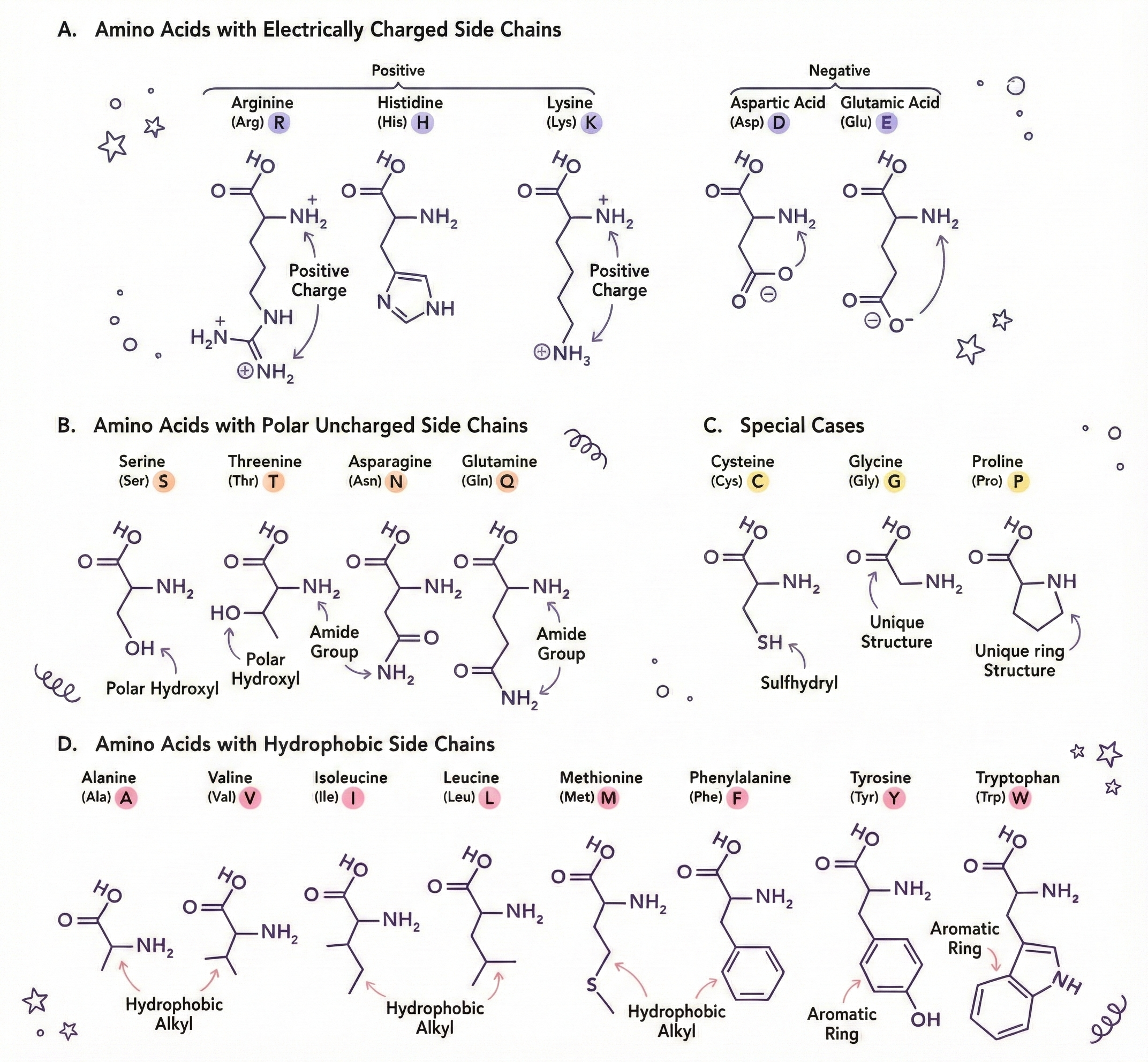
Let Me Introduce You to Some Amino Acids (They Have Personalities)
- Glycine – smallest; side chain is just a hydrogen atom. Flexible and a great team player.
- Proline – a ring structure that creates kinks; the rebel that forces bends.
- Cysteine – contains sulfur; two cysteines can form a disulfide bond (S‑S), acting like a chemical staple.
- Tryptophan – large and bulky; typically buried in protein cores because it’s hydrophobic.
- Aspartic acid & Glutamic acid – negatively charged; repel other negatives and attract positives.
- Lysine & Arginine – positively charged; attract negative groups and stabilize structures.
- Phenylalanine, Leucine, Isoleucine, Valine – hydrophobic; they cluster together away from water.
The side chains dictate:
- Hydrophilicity vs. hydrophobicity
- Charge (positive, negative, neutral)
- Size (affects packing)
- Rigidity vs. flexibility
- Chemical reactivity
The Combinatorial Explosion of Possibilities
Typical proteins contain 200–400 amino acids; some, like titin, have 34,350.
For a modest 100‑amino‑acid protein, the number of possible sequences is 20¹⁰⁰ ≈ 1.27 × 10¹³⁰.
- Observable universe atoms: ~10⁸⁰
- Observable universe stars: ~10²⁴
Thus, possible 100‑residue sequences outnumber atoms in the universe by roughly 10⁵⁰. Most of these sequences never fold into functional structures; they aggregate or are degraded. Evolution, over 3.5 billion years, performed a massive brute‑force search to find the few viable folds.
We lack that timescale, so we need smarter methods to design proteins now.
Folding, The Part Where the Magic Happens (And Also Where Everything Can Go Wrong)
When a ribosome finishes translating a protein, it releases a long, linear polypeptide chain. The backbone consists of repeating NH‑CHR‑CO units linked by peptide bonds:
...—NH—CHR—CO—NH—CHR—CO—NH—CHR—CO—...Side chains (R groups) protrude from this backbone. Immediately after synthesis, the chain begins to explore conformations, seeking the energetically favorable 3D structure dictated by its sequence. This folding process is rapid, highly cooperative, and essential for function—yet prone to errors that can lead to disease.